Researchcp
Overview
The research conducted by this laboratory spans a wide range of topics in genomics, molecular biology, and computational biology, showcasing a multidisciplinary approach to understanding fundamental biological processes. The team has made significant contributions to the field of immunology, particularly in the realm of T-cell receptor (TCR) and epitope interactions. Pioneering work includes the development of advanced computational models such as ATM-TCR, a multi-head self-attention model for predicting TCR-epitope binding affinities, and PiTE, a TCR-epitope binding affinity prediction pipeline utilizing a Transformer-based Sequence Encoder. These computational tools provide valuable insights into the dynamics of immune responses and have potential applications in vaccine development and personalized medicine.
Moreover, the research team delves into the intricacies of DNA replication and transcription processes, shedding light on the factors influencing mutation rates and transcript errors in bacterial pathogens like Salmonella enterica subsp. enterica and Escherichia coli. Their work not only uncovers the complexities of mutagenesis but also challenges existing paradigms, as seen in studies on DNA replication-transcription conflicts and the symmetrical wave pattern of base-pair substitution rates across the E. coli chromosome. Beyond the laboratory bench, the team engages with broader societal issues, addressing privacy and ethical considerations in wastewater monitoring. Their commitment to understanding and advancing both the theoretical and practical aspects of genomics and molecular biology positions this research group at the forefront of innovative scientific inquiry.
Active Learning Framework for Cost-Effective TCR-Epitope Binding Affinity Prediction
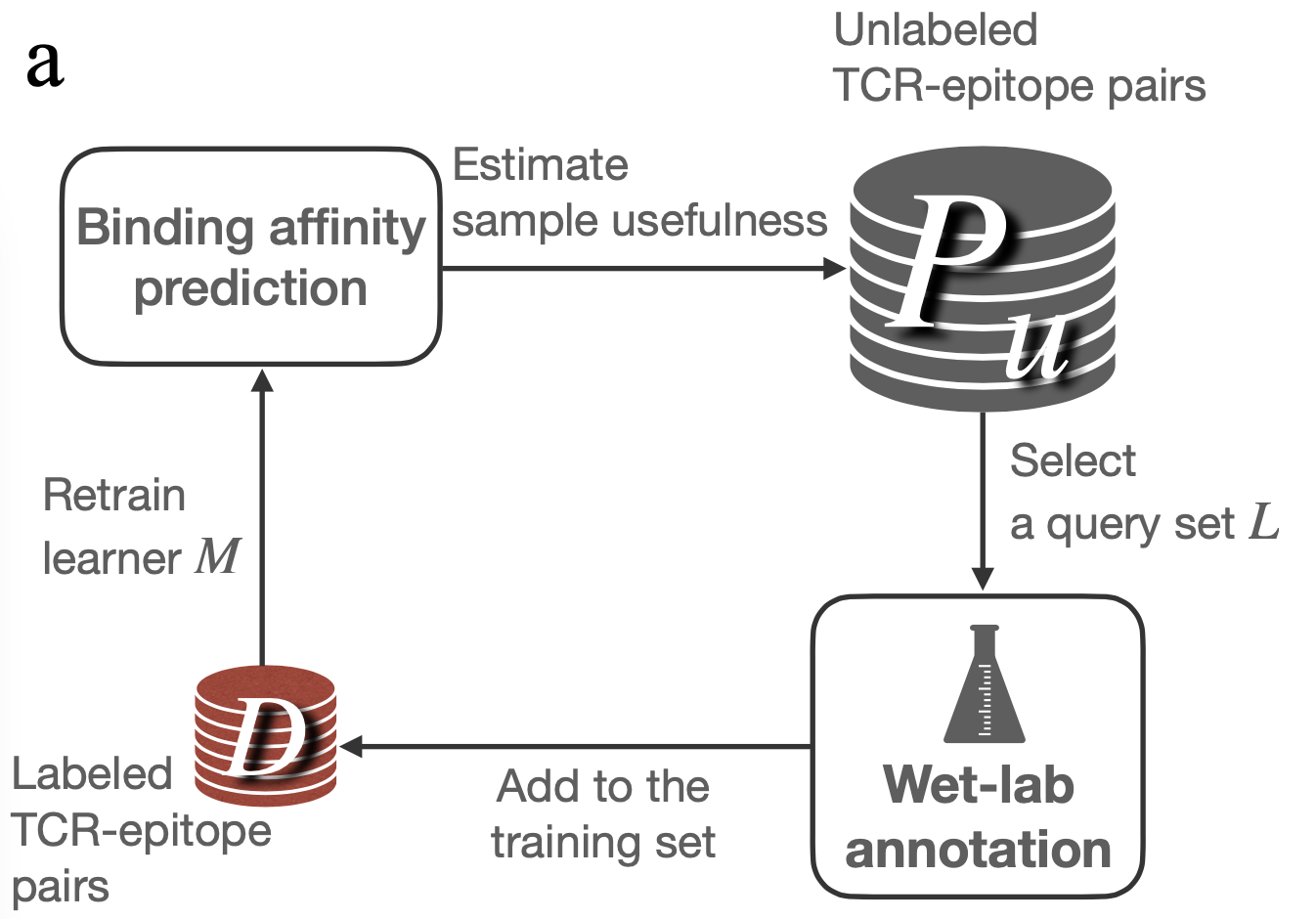
Description:
A unified data optimization framework (dubbed ActiveTCR) that integrates active learning and TCR-epitope binding affinity prediction models. In two distinct use cases, ActiveTCR demonstrated superior performance over passive learning, notably cutting annotation costs approximately half and minimizing redundancy by over 40% - all without compromising on model performance. ActiveTCR stands as the first systematic exploration into the realm of data optimization for TCR-epitope binding affinity prediction.
Publication(s):
https://github.com/Lee-CBG/ActiveTCR
Team members:
Pengfei Zhang, Seojin Bang, Heewook Lee
Context-Aware Amino Acid Embedding Advances Analysis of TCR-Epitope Interactions

Description:
Introducing catELMo, a groundbreaking and efficient amino acid embedding model, specifically tailored for T cell receptors. This advanced model facilitates an impressive boost of over 20% in absolute AUC when predicting binding affinity for unseen or novel epitopes, outperforming the conventional BLOSUM62. Moreover, catELMo exhibits an extraordinary capacity to maintain comparable performance to BLOSUM62, while reducing about 93% training data, making it a game-changer in the field of TCR analysis.
Publication(s):
https://elifesciences.org/reviewed-preprints/88837v1
Team members:
Pengfei Zhang, Seojin Bang, Michael Cai, Heewook Lee
PiTE: TCR-epitope Binding Affinity Prediction Pipeline using Transformer-based Sequence Encoder
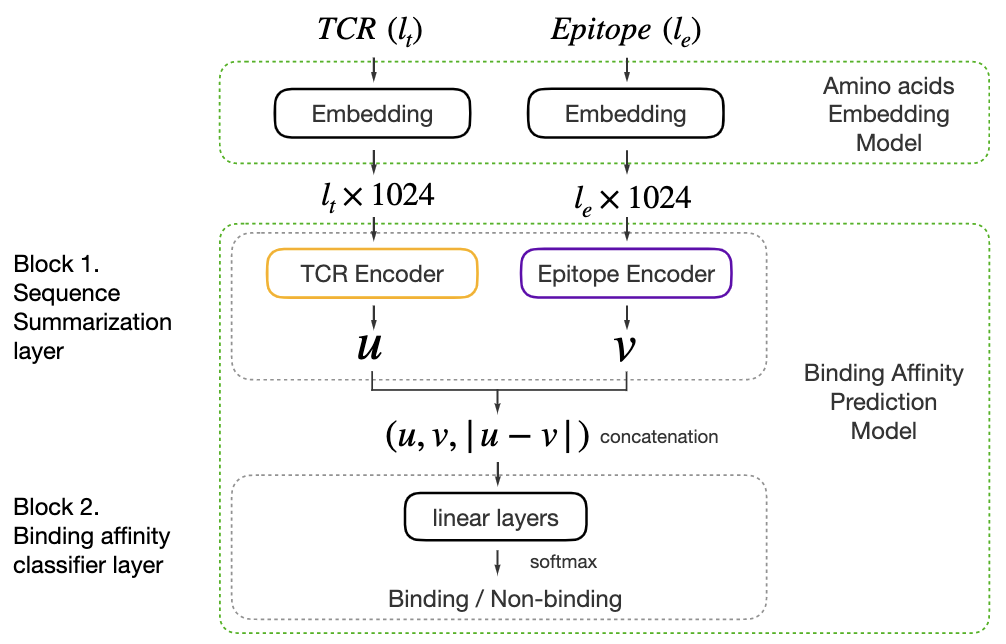
Description:
How to better summarize multiple amino-acid-level embeddings into a single sequence-level embedding compared to average pooling? We build sequence encoders utilizing various structures including Transformer, BiLSTM, and ByteNet, and propose PiTE, a state-of-the-art two-step pipeline designed for TCR-epitope binding affinity prediction..
Publication(s):
https://www.worldscientific.com/doi/pdf/10.1142/9789811270611_0032
Team members:
Pengfei Zhang, Seojin Bang, Heewook Lee
ATM-TCR: TCR-Epitope Binding Affinity Prediction using a Multi-Head Self-Attention Model
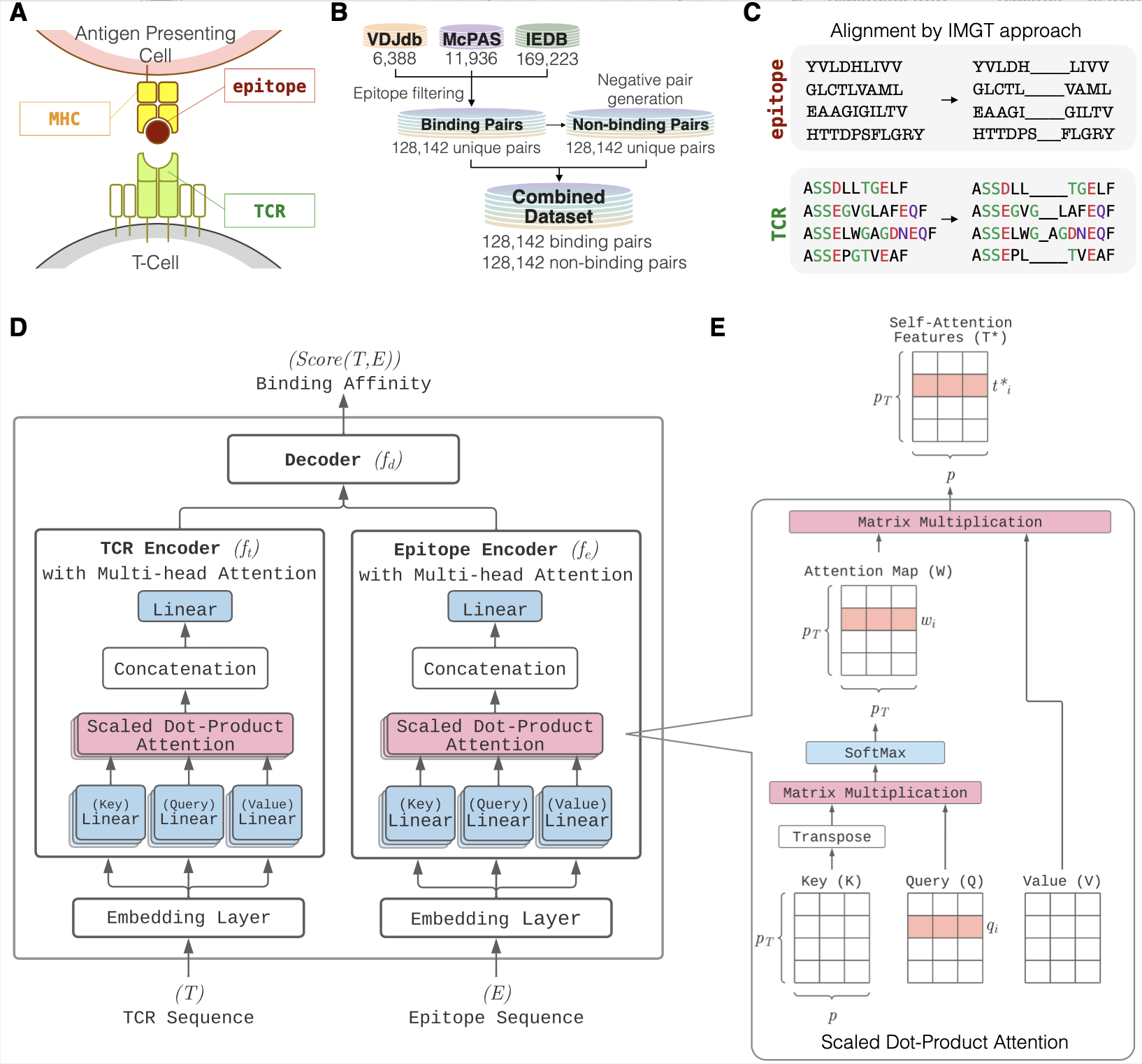
Description:
ATM-TCR leverages multi-head self-attention mechanisms to capture biological contextual information and improves generalization ff TCR-epitope binding affinity prediction models. A novel application of the attention map to improve out-of-sample performance by demonstrating on recent SARS-CoV-2 data.
Publication(s):
https://www.frontiersin.org/articles/10.3389/fimmu.2022.893247/full
Team members:
Michael Cai, Seojin Bang, Pengfei Zhang, Heewook Lee
